Microsoft word - informe def iii cevime_paliperidona.doc
COMITÉ DE EVALUACIÓN DE NUEVOS MEDICAMENTOS DE EUSKADI COMITÉ DE EVALUACIÓN DE NUEVOS MEDICAMENTOS DE EUSKADI INFORME DE EVALUACIÓN PALIPERIDONA Comparador: HALOPERIDOL, ANTIPSICÓTICOS ATÍPICOS NO SUPONE UN AVANCE TERAPÉUTICO. Nombre Comercial y presentaciones: INVEGA® (Janssen-Cilag) 3 mg 28 comprimidos de liberación prolongada (140,75 €) 6 mg 28 comprimidos de
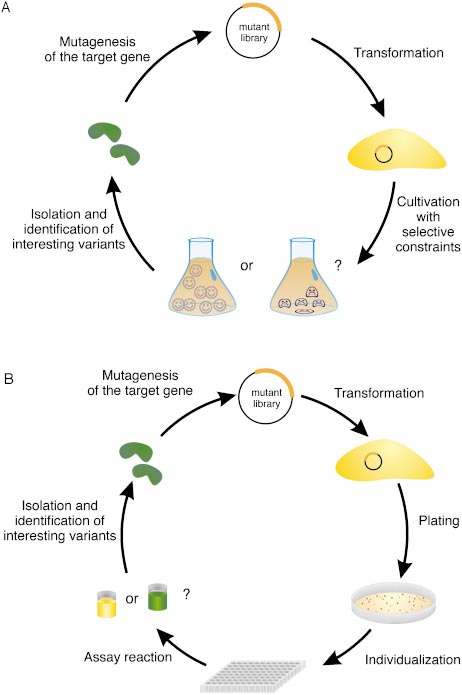 extremely rare. Codon-level random mutagenesis ofcomplete genes therefore would be desirable, but hasnot been realized yet.
extremely rare. Codon-level random mutagenesis ofcomplete genes therefore would be desirable, but hasnot been realized yet.