septodont.at
Fachinformation Bezeichnung des Arzneimittels 2. Qualitative quantitative Zusammensetzung Die vollständige Auflistung der sonstigen Bestandteile siehe Abschnitt 6.1. Darreichungsform Sprühlösung zur Lokalanästhesie, Pumpspray 4. Klinische 4.1 Anwendungsgebiete - Oberflächliche Betäubung und Desinfektion der Mundschleimhaut vor einer - Oberflächliche Betäu

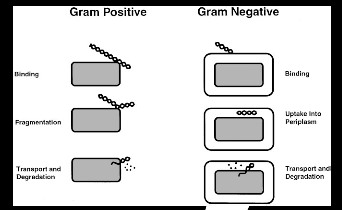 Figure 4.1: Transformation pathways in gram-positive and gram-negative
Figure 4.1: Transformation pathways in gram-positive and gram-negative 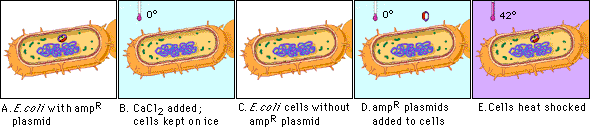 This method works well for circular plasmid DNAs but not for linear molecules such as fragments of chromosomal DNA. Figure 4.2: Overview of competence and heat shock
From: http://www.phschool.com/science/biology_place/labbench/lab6/test1.html
This method works well for circular plasmid DNAs but not for linear molecules such as fragments of chromosomal DNA. Figure 4.2: Overview of competence and heat shock
From: http://www.phschool.com/science/biology_place/labbench/lab6/test1.html